Help :
piRSNP is a database providing comprehensive resources of piRNA-related single nucleotide polymorphisms (SNPs) of human and mouse. Users could freely view it for providing a valuable resource for piRNAs research and application.
We comprehensively identified SNPs on piRNAs by examining the loci of both SNPs and piRNAs coordinates and the workflow is illustrated in Figure 1 (a). 42,967,522 SNPs on 10,773,081 human piRNAs are identified. The SNP density of each chromosome is shown in Figure 2 and drew by RIdeogram. In piRSNP, there was an average of 1,790,312 SNPs for each chromosome, ranging from 7,883,268 for chromosome 4 to 33,807 for chromosome Y.
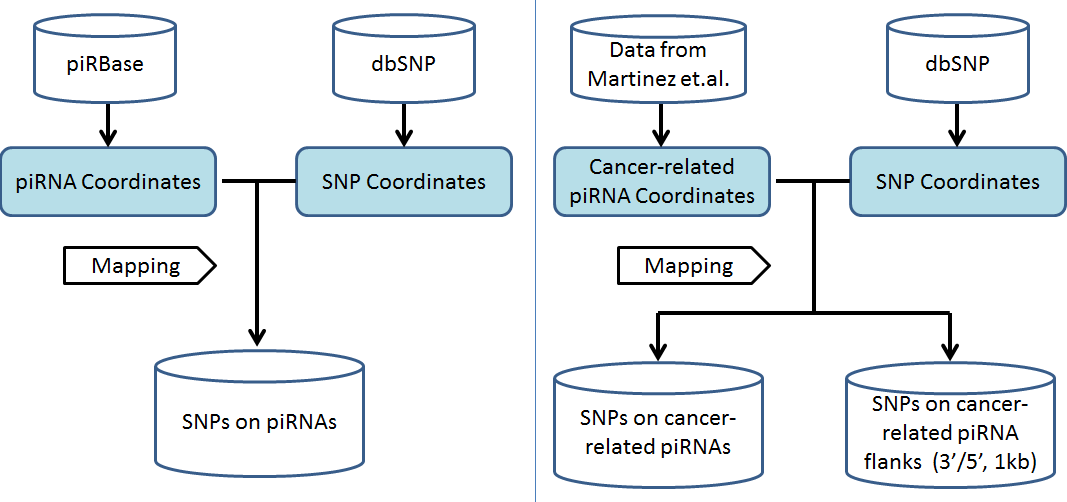
Figure 1 Workflow depicting analysis of SNPs on piRNAs.
We identified SNPs located both in piRNAs and in their adjacent 1kb upstream and downstream regions by mapping the coordinates.
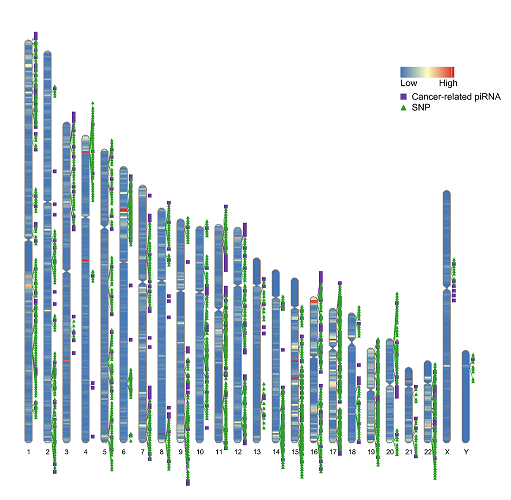
Figure 2 SNPs on cancer-related piRNAs and piRNAs distribution across the whole human genome
Note: The overlaid heatmap presents the density of the SNPs on piRNAs, the purple boxes labels cancer-related piRNAs locus and the green triangles labels SNPs locus in cancer-related piRNAs across the human genome. Annotation information was downloaded from the GENCODE website (https://www.gencodegenes.org).
Two tools, namely miRanda and BLAST, are used for predicting cancer-related piRNAs’ targets.
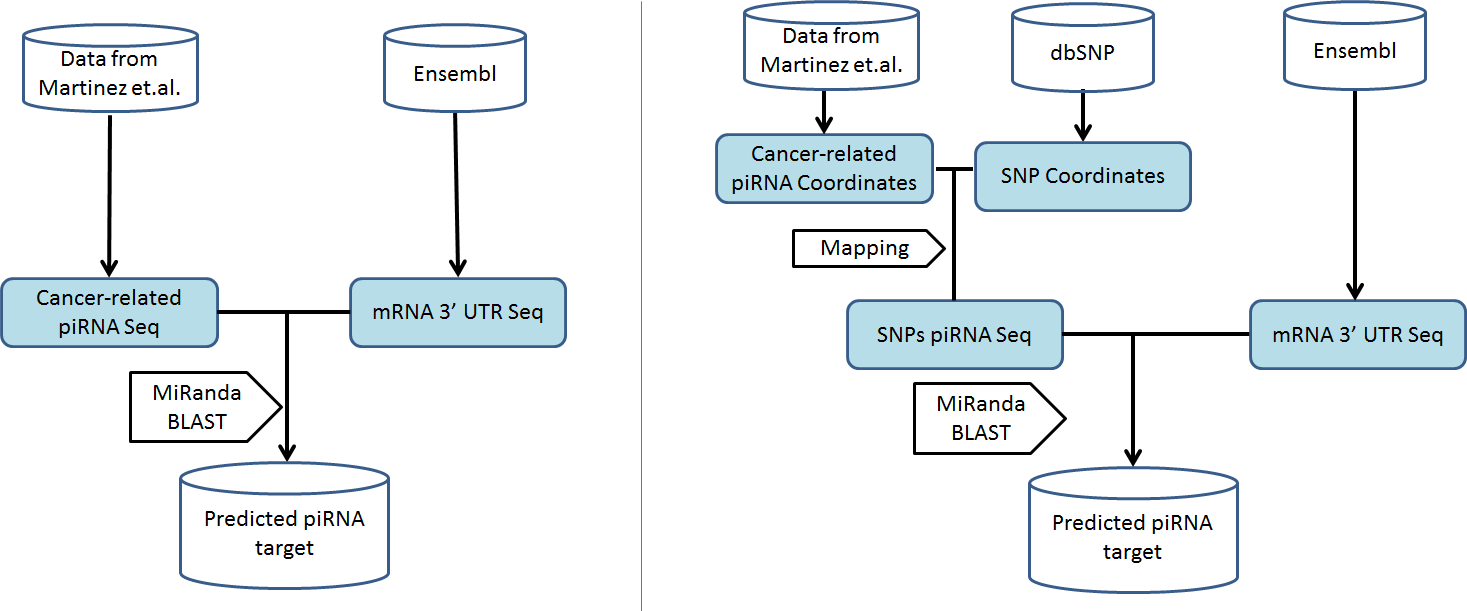
Figure3 Workflow depicting analysis of target about piRNAs and piRNAs with SNPs
